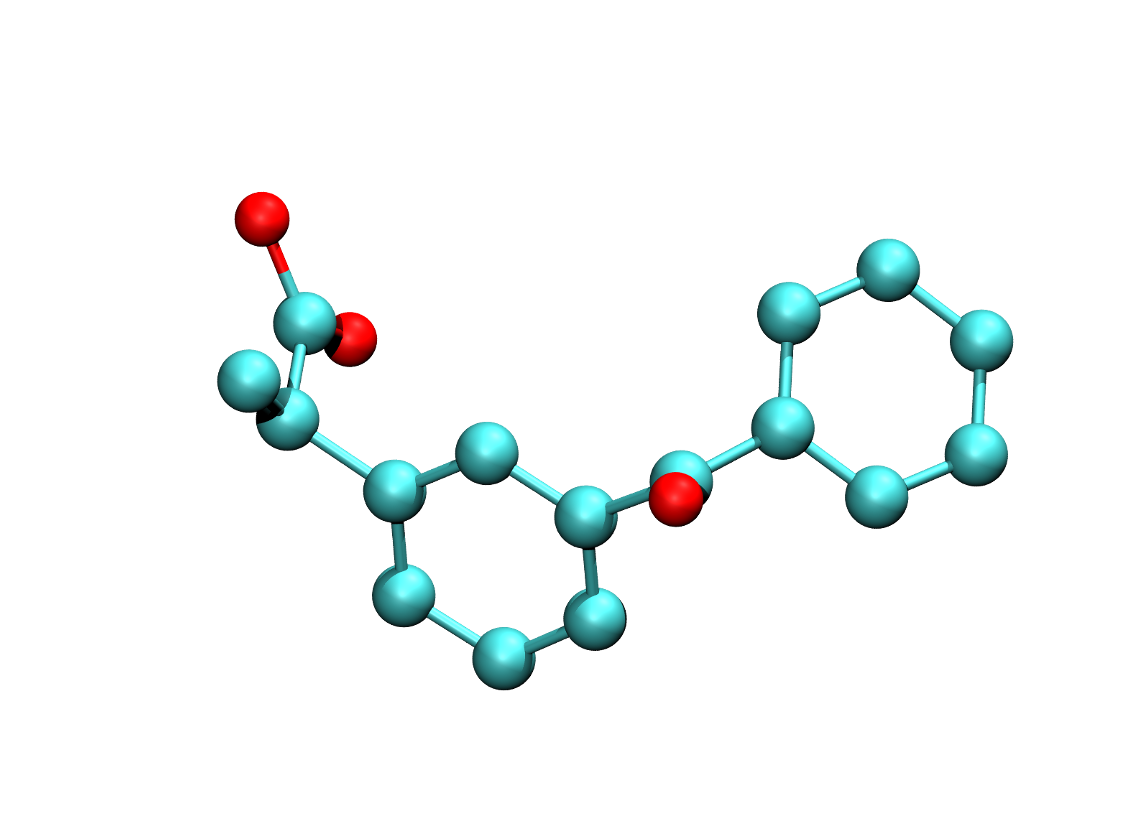
Molecular Parameter Generation
A module to generate the forcefield parameter of organic molecules.
- Support structure formats of SMILES, .mol2, .mol, .pdb, .gjf, .xyz, .cif, and more.
- Support GAFF/GAFF2 forcefield.
- Calculate the RESP or AM1-BCC charge in several minutes.
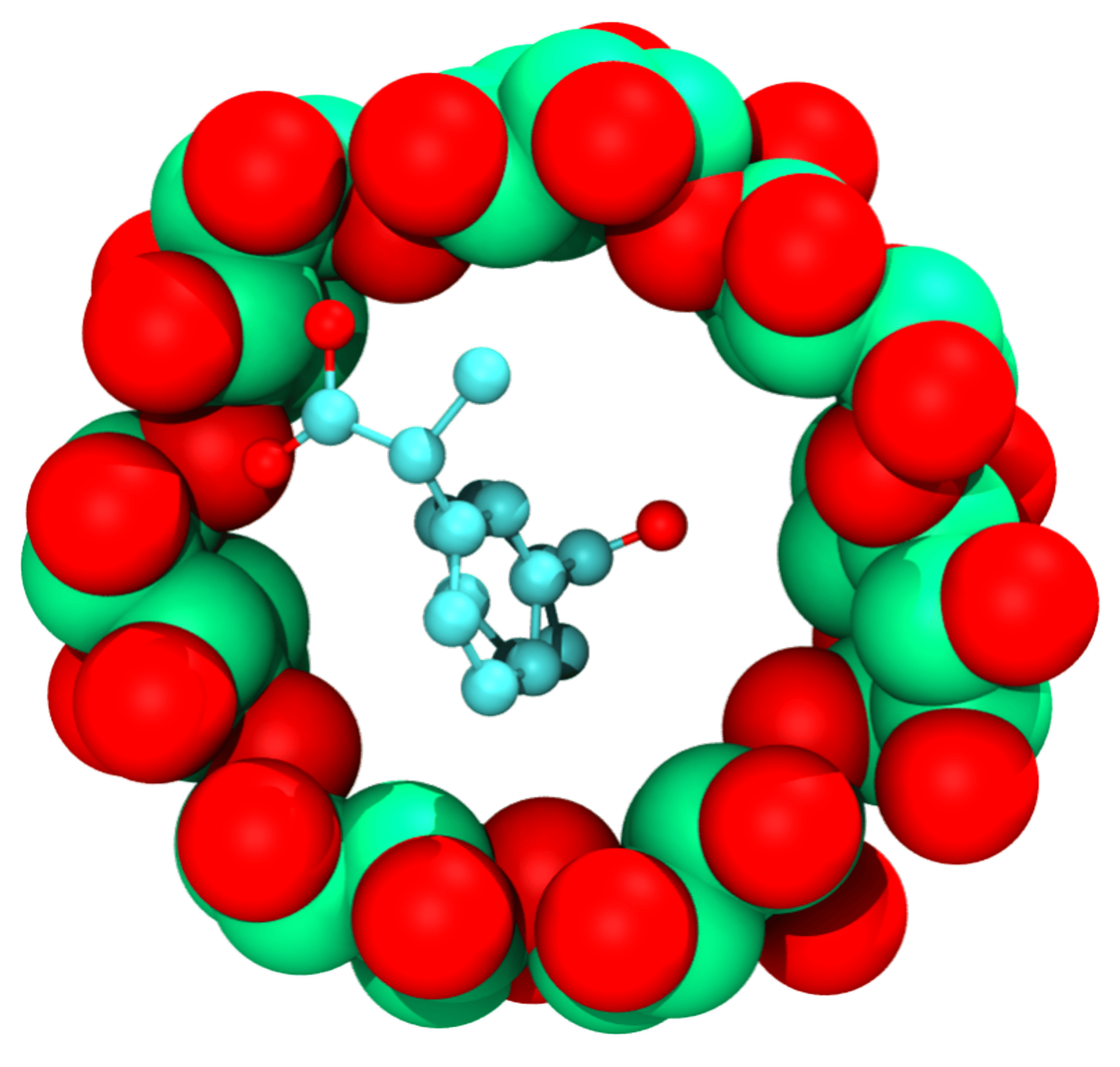
Cylodextrin
A module to investigate the binding property of drug and cyclodextrin.
- Generate the binding pose between drug and cyclodextrin based on molecular docking.
- Generate the initial file including topology and structure for molecular dynamics simulation of cyclodextrin-drug complex.
- Support the Molecular dynamics simulation and Customized result analysis.
- Support the MM-PBSA bindging free energy calculation.

Micelle
A module to investigate the molecular mechanism of drug and micelle.
- Generate the initial file including topology and structure for molecular dynamics simulation of drug and micelle.
- Support the Molecular dynamics simulation and Customized result analysis.
- Support the Self-assembly and Preformed model methods.
- Support the Coarsed-grained and All-atom forcefields.
- Support the three morphology including Sphere, Cylinder and Capped-cylinder.

Solid dispersion
A module to invesigate the molecular mechanism of drug and dispersion.
- Generate the initial file including topology and structure for molecular dynamics simulation of drug and polymer.
- Support the Molecular dynamics simulation and Customized result analysis.
- Support two formulation manufacture methods including High temperature melting and Solvent removing.
- Support the common polymer excipient forcefield.

Liposome
A module to investigate the molecular mechanism of drug and liposome.
- Generate the initial file including topology and structure for molecular dynamics simulation of drug and liposome.
- Support the Molecular dynamics simulation and Customized result analysis.
- Support the Self-assembly and Preformed model methods.
- Support the Classical, Pegylated and Protein-modified liposome .
- Support the Coarse-grained model based on MARTINI forcefield.
- Support the MARTINI forcefield parameter generation of drug.
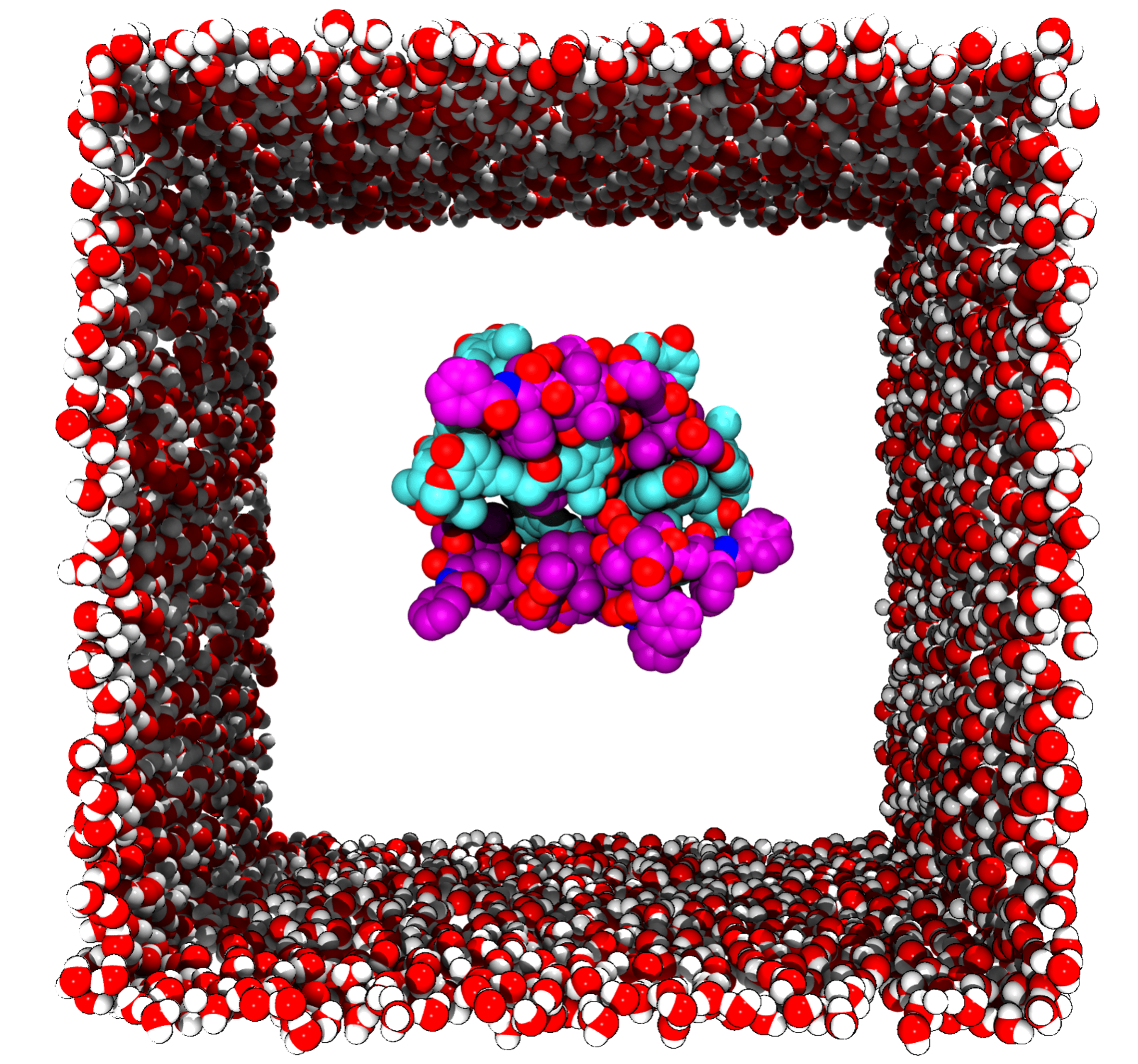
Self-assembly drug nanoparticle
A module to investigate the self-assembly process of drug nanoparticle.
- Generate the initial file including topology and structure for molecular dynamics simulation of drug and excipient.
- Support the Molecular dynamics simulation and Customized result analysis.
- Support the multiple input of drug molecules.
- Support the Excipient input.

Drug transmembrane delivery
A module to investigate the transmembrane process of drug molecules.
- Generate the initial file including topology and structure for molecular dynamics simulation of drug and membrane.
- Support the Molecular dynamics simulation and Customized result analysis.
- Support the Lipid17 forcefield.
- Support the Umbrella sampling.